J-RGC
JAM-B cells (also known as J-RGCs) are a molecularly defined subcategory of direction-selective retinal ganglion cell found in mice. Their deepest importance to the study and categorization of cells in the retina lies in that they were the first subtype of retinal ganglion cell to be classified based on the presence of a molecular marker.
JAM-B cells are named after Junctional Adhesion Molecule B (JAM-B), the molecular marker which defines them as a distinct subset of retinal ganglion cells. JAM-B cells share many functional and structural traits in addition to the presence of this molecule. Most J-RGCs have asymmetric dendritic arbors aligned in a dorsal-to-ventral direction across the retina. Those J-RGCs with markedly asymmetric dendritic arbors also have asymmetric receptive fields.
In addition, J-RGCs are direction-selective, responding specifically to upward motion. Furthermore, they are all OFF RGCs, meaning they respond only to the trailing edge of a bright stimulus [1]
Contents
Anatomy
Shape
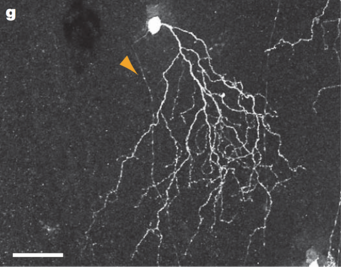
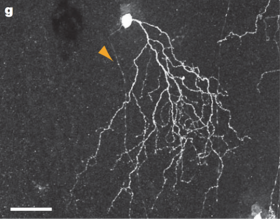
In about 85% of JAM-B cells, there exists a marked asymmetry in the dendritic arbor. More than 90 percent of the dendritic arbor lies on one side of the soma. Asymmetry of the dendritic arbor is an unusual feature for a Retinal Ganglion Cell, as the dendrites of most Retinal Ganglion Cells are symmetrical. Therefore, the prevalence of this asymmetry in such a high proportion of JAM-B cells is notable. Even more notable is that the asymmetrical dendrites of JAM-B cells all point in the same direction, about 13 degrees nasal of ventral.
Alternatively, approximately 15% of JAM-B cells are not markedly asymmetrical. These non-asymmetrical JAM-B cells are located near the dorsal and ventral margins of the retina. Therefore, nearly all JAM-B cells in the nasal, central, and temporal regions of the retina are markedly asymmetrical [1]
Connections
The dendrites of JAM-B cells have been found to arborize in a narrow band between processes of dopaminergic and cholinergic amacrines Indeed, the dendrites of both symmetric and asymmetric JAM-B cells are confined to the outer third of the inner plexiform layer. [1]
Kim et al. used yellow fluorescent protein to trace the projection of JAM-B cells to the mouse brain. They found that JAM-B cells project heavily to the superior colliculus. Thus it seems that mice have invested heavily in the detection of upward motion. It remains to be determined why exactly this is the case.
Physiology
JAM-B cells respond selectively to upward motion. It has been proposed that the asymmetry of a JAM-B cell's dendritic arbors is related to this direction selectivity. Kim et al. state, "across the entire J-RGC population, the degree of direction selectivity was correlated with the degree of asymmetry of the receptive field (Supplementary Fig. 6). Thus, within a single molecularly defined class of OFF-RGCs, dendritic structure and cell function are closely linked, suggesting that the latter arises from the former." [1]
Whereas the selectivity of other direction-selective RGCs depends on input from starburst amacrine cells, which themselves show directional responses, J-RGCs receive little input from these cells and thus must rely on other mechanisms. One possible mechanism is suggested by the finding that inhibitory synapses on some RGCs are concentrated at distal dendrites. Distal inhibition on the asymmetric dendrites of J-RGCs could account for their asymmetrically displaced surround. (Previous two sentences taken directly from Kim et al. paper)
The receptive field of J-RGCs is unusual in that the integrated strength of its ON surround exceeds that of the OFF centre. (Kim et al.) Studies imply that no sizable populations of OFF RGCs exist with asymmetric dendrites pointing in directions other than dorsal-to-ventral (source: Kim et al.)
Molecules
JAM-B Cells are classified by the expression of the Junctional adhesion molecule B (JAM-B) protein.
Junctional adhesion molecule B is a protein that in humans is encoded by the JAM2 gene. JAM2 has also been designated as CD322 (cluster of differentiation 322). Tight junctions represent one mode of cell-to-cell adhesion in epithelial or endothelial cell sheets, forming continuous seals around cells and serving as a physical barrier to prevent solutes and water from passing freely through the paracellular space. The protein encoded by this immunoglobulin superfamily gene member is localized in the tight junctions between high endothelial cells. It acts as an adhesive ligand for interacting with a variety of immune cell types and may play a role in lymphocyte homing to secondary lymphoid organs [2].
Development
History
JAM-B cells were first discovered by In-Jung Kim, Yifeng Zhang, Masahito Yamagata, Markus Meister, and Joshua R. Sanes of Harvard's Department of Molecular and Cellular Biology and Center for Brain Science.
To mark JAM-B cells for structural and functional study, they generated mice that expressed a ligand–activated Cre recombinase oestrogen receptor fusion protein 10 (CreER) under the control of regulatory elements from the JAM-B gene.
Open Questions/Status
Relationship between J-RGCs and direction-selective collicular cells
The collicular termination of J-RGCs (of which JAM-B cells are a majority) is intriguing in light of a study in which Dra"ger and Hubel mapped the receptive fields of neurons in the superior colliculus of the mouse. Nearly all of the direction-selective neurons they studied (35 out of 38) preferred upward motion in the visual field. This preference corresponds to that of J-RGCs [1]. Kim et al. have proposed that the receptive fields of direction-selective collicular cells are built from J-RGCs. Research remains to be done on this topic.
Sensitivity to upward motion in mice
It is uncertain why the mouse seems to have invested so heavily in sensitivity to upward motion. KIm et al. have suggested mating JAM-B–CreER mice to other transgenics bearing appropriate Cre-activated channels or toxins. Thus it could be possible to inactivate the pathway and thereby directly test its function[1].
Eyewire progress
|
File:Jamb010 top.png 280px |
File:Jamb010 phys.png 140px |